HARP - A Web Resource for Predicted Structural Impacts of Anti-microbial Resistance Mutations in Leprosy
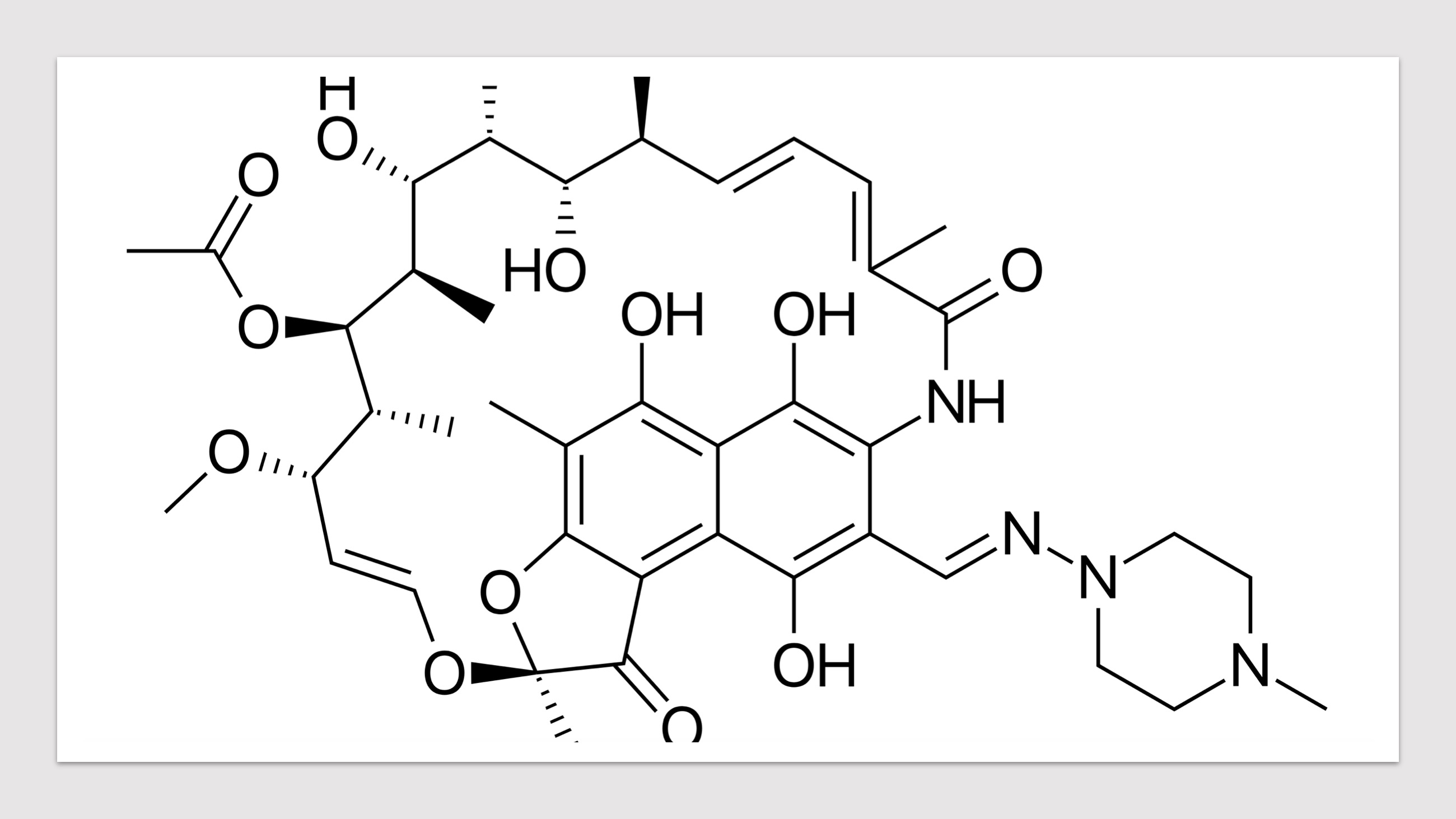
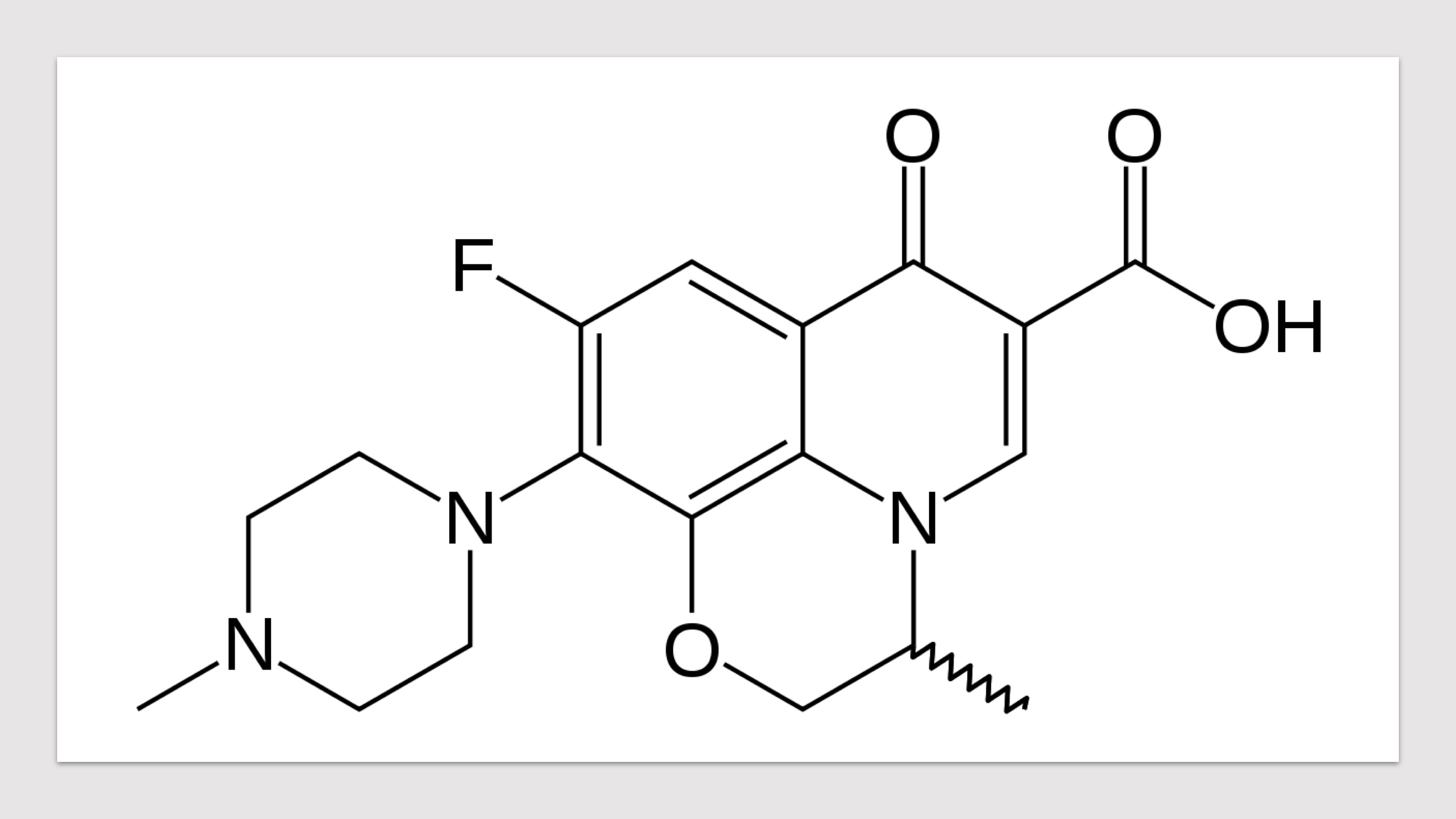
HARP (a database of Hansen's Disease Antimicrobial Resistance Profiles) is an integrated web-resource with predicted structural implications of systematic mis-sense mutations in known drug targets of Mycobacterium leprae. A web database of computationally inferred sequence and structure based predictions of stability and vibrational entropy changes due to mutations. A resource to inform the impacts of known and emerging mutations on protein-protein, protein-ligand and protein-nucleic acid affinities, and consequent anti-microbial resistance outcomes in leprosy. HARP is developed with the aim of supporting mycobacterial research community with essential knowledge on the structural implications of mutations conferring antimicrobial resistance in leprosy.
Predicted Impacts of Systematic Missense Mutations in Drug Targets
- Note:
- Click the buttons with the drug names to query mutations in corresponding drug targets.
- Click on the pictures to query corresponding drugs in PubChem.
Disclaimer: This resource is intended for purely research purposes.
Citation: Vedithi SC, Malhotra S, Skwark MJ, Munir A, Acebrón-García-De-Eulate M, Waman VP, Alsulami A, Ascher DB, Blundell TL. HARP: a database of structural impacts of systematic missense mutations in drug targets of Mycobacterium leprae. Computational and Structural Biotechnology Journal. 2020 Nov 19, doi: https://doi.org/10.1016/j.csbj.2020.11.013